Emmanuel Vega, Life Science
Abstract
Bacteriophages offer a solution to antibiotic resistance through research and implementation. This project explores and compares the treatment of Bacteria (Escherichia coli K12) with bacteriophages. The purpose of the project is to investigate whether bacteriophages can stimulate mutation in E. coli K12. This is investigated by exposing E. coli K12 to a gradient of concentrations in an 8 by 8-inch plate. This was done to pursue a unique insight into microbial adaptation and visualization of evolutionary dynamics while promoting the usage and innovation of bacteriophage.
Introduction
Antibiotics have been available since the 1930s and have saved countless lives, increasing the average life expectancy from 47 to 78 years (Adedeji 2016). However, antibiotics are being massively overused. (Brown 2015) Antibiotic overuse causes antibiotic resistance, threatening international health because bacterial strains evolve and render antibiotic treatment ineffective. The strain Staphylococcus aureus is an example of a fully resistant strain to methicillin. It is one of the most common antibiotic-resistant bacteria which causes life-threatening bloodstream and surgical-site infection (Longitude management team 2014). The increase in antibiotic resistance highlights the importance of improving the way we treat bacterial infections. Bacteriophages (phages) are viruses that target bacteria. In addition, they are specific to bacteria species and are harmless to eukaryotic cells. (Kasman 2022)
Therefore, a type of bacteria such as T4 t coli species only. The ability to being able to target a specific bacterium opens the utility of the phages in the medical world. If an individual was infected with a dangerous strain of bacteria such as the flesh-eating bacteria Staphylococcus aureus. (Pincus 2015) The patient could undergo phage therapy to kill the bacteria.
Bacteriophages have seen great innovation in recent times. Phage therapy is in the emerging steps and there are case studies of phage therapy saving lives and treating dangerous antibiotic-resistant strains. This instance occurred when Tom Patterson, an elderly gentleman contracted the bacteria Acinetobacter baumannii, while on his trip to the middle east. After physicians found that antibiotics were not functioning, they decided to attempt to use bacteriophage therapy (Scott LaFee et al 2017). There are many trials including the trial where 20 patients with cystic fibrosis and multidrug resistant mycobacterium were tested. (Dedrick 2023)
This project will see to what degree bacteria mutate when exposed to phages according to a gradient of phage concentrations. This is to explore how the mutation spread of bacteria is affected by exposure to a gradient of bacteriophages.
Materials and Methods
T4 coli Phage Proliferation
This experiment needs many bacteriophages. Therefore, techniques like a plaque assay were used to proliferate phages. This was done by making a solution of liquid agar, and T4 Phages and placing them in a Petri dish. The step was done by filling a test tube with liquid agar, then adding 1000 uL of phage broth into the mixture. The agar and phage mixture were well mixed and then placed in a sterile petri dish. The Petri dish was shaken well to spread the solution over the plate. The petri dish was closed and left on the side to solidify while the other 6 Petri dishes were prepared in the same manner. Once the 6 plates were prepared, they were placed in the incubator for 30 minutes. The remaining phages were kept in the fridge for storage. Acquiring an agar bottle and melt it in the microwave. The bottle was set aside to cool down in cold water. The agar bottle was then placed in a hot water bath heated at 45*C. A temperature gun was used to check that the liquid agar was at the correct temperature then, the agar was poured as an agar overlay on the plates with the mixture of agar and phages. Then, the plates were left by flame to solidify and then we inversely placed them in the incubator.
Phage extraction
After incubating the plates with phages. Plaques were observed. A small circular hole-like structure at the bottom of the plate under the agar overlay. Subsequently, a sterile p200 pipette tip was taken, gently stabbed the top agar layer in the center of the plaque while avoiding the nearby bacteria around. Then a centrifuge tube was filled with 500uL of phage buffer using a p1000 and subsequently placed the pipette tip with the plaque in a centrifuge tube filled with 500uL of phage buffer. The centrifuge tube was closed and shaken, and the mixture was transferred to a 500uL beaker. The beaker was filled prefilled with 50mL of phage buffer and 10mL of T4 phages from the stock solution. The plaques were repeatedly picked and placed in the beaker until they measured 250mL. This part was a lengthy process and took 2 hours to complete. The phage solution was stored in the fridge.
Making phages
To make the phage mixtures, a microwave was first used to melt agar, then it was cooled it by putting it in a cold-water bath and before it solidified. Subsequently it was placed in a hot water bath at 45-degree temperature to keep it liquid. However, 45 degrees is also an adequate temperature for the phages to activate, and it is a temperature where the antibiotic will not break down. Once the agar was ready, it was carefully poured according to the wanted percent concentration and added the corresponding percent product. Finally, the mixing step was repeated while changing the concentration of the product.
Pouring the agar mixtures
Agar was melted and cooled while regulating its temperature in a water bath. Different solutions in a 50ml sterile centrifuge tube were mixed. Two tubes were used to total 100ml. The solution was well mixed by swirling the beaker and carefully pouring it onto the 8 by 8-inch Pyrex tray. Starting first with 0x solution and proceeding with the subsequent mixtures. A Solid plastic sheet was covered with aluminum and separated by clean saran wrap and a plastic which was cut from a plastic cutting board and wrapped in aluminum foil; it was also sterilized. The saran wrapped separate sections and prevented leaks. The plastic sheet is used to group the agar solutions. The agar started to solidify and then the steps were repeated including the pouring step but with the subsequent increasing tenfold concentration until all mixtures were done and 5 bands of agar with a different amount of product were available.
Spreading bacteria
A centrifuge (1mL) tube was filled with distilled water and using a pipette tip small number of bacteria was scraped and placed it in a centrifuge tube. The tube was shaken to spread the bacteria, subsequently a p200 pipette was taken to pipette drops of the bacteria/water mixture in the control section (0x concentration). An L spreader was used to evenly spread the bacteria across the area. The step was repeated, however the bacteria was applied across the edge of the Pyrex tray moving up the gradient of concentrations to set up a positive control
Incubator
The Pyrex tray with the agar mixtures and bacteria were placed into the incubator at 25 degrees Celsius.
Data collection
The growth of bacteria was observed over a week and a half (around 10 days). A phone camera was used to record and take images of the growth and death of bacteria. The size of mutants as they traversed the dish was measured using a precise ruler. Lastly the percent cover of plaques in each band and overall plate was calculated in addition to counting Plaque forming units.
Results
In this first trial it was difficult to see results, therefore the results could not be used to support or refute the null or alternative hypothesis.
So, models were made on Bio render to predict the outcome of our future tests. Figures 4 and 5 show the null hypothesis and potential results. Figures 6, 7, and 8 show the work done and the trials still being conducted to enhance the experiment.
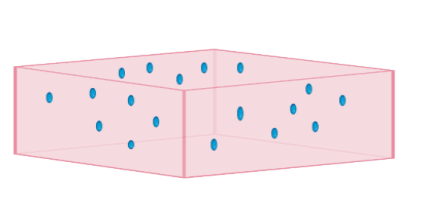
Figure 1. How phages are spread out on the agar. Phages are spread out in random places, not all sections of the agar necessarily contain. This Bio Render model provides a base understanding for future experiments.
Orange = plate with agar
Green = Bacteria
Circle = Plaque
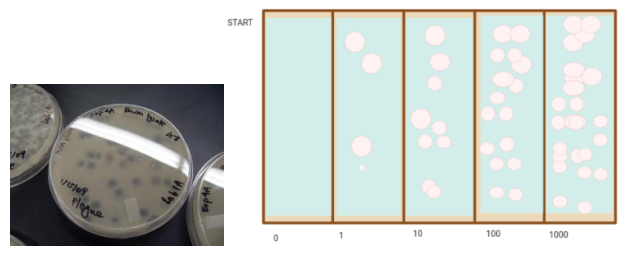
Figure 2. The results. This is based on plaque assay results, and it was designed on Bio Render.

Figure 3. The plate set up. E. coli K12 in MEGA plate. Bacteria are too small to see. We must maintain a safe distance from the bacteria with our camera to prevent contamination and ensure safety therefore closer photos cannot be taken.
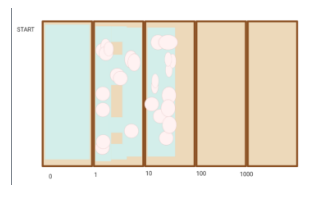
Figure 4. demonstrates our expected outcome of Null Hypothesis modeled on biorender: Gradient inhibiting mutating of bacteria. The outcome sees high percentage cover of plaques/zones of inhibition. And does not reach higher concentrations
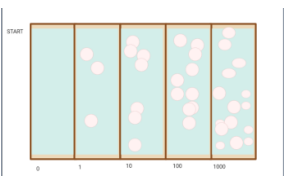
Figure 5. Expected outcome of our Alternative Hypothesis: Gradient stimulates bacterial mutation and resistance. Outcome sees high growth and spread of bacteria across the entire surface. Percentage cover of plaques/zones of inhibition is seen but bacteria spreads all over surface.
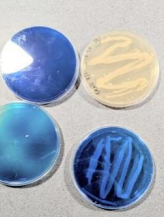
Figure 6. How we are optimizing methods for the next test: blue plates for greater contrast.

Figure 7. Lab set up
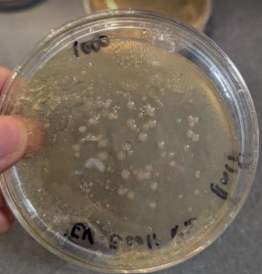
Figure 8. The preliminary test: Phage proliferation
A plaque assay was done after the experiment to demonstrate that higher concentrations of phages does kill more bacteria, this is
Discussion
Interpreting Results
The procedure was followed and focused only on bacteriophages as it is of vast importance to explore the field and all factors of the bacteriophage so we can use it to save many lives. This experiment was followed correctly, and efforts were made to minimize errors. However, results could not be conveyed due to the lack of visibility of plaques and small, undistinguishable zones of inhibition. The plate was able to see zone some zones where bacteria did not grow if one looks closely however, the information does not give ground to accept/reject the null/alternative hypothesis. The small amount of growth can be attributed to the lack of growth of E. coli. Which can be linked to factors such as temperature and phage distribution. Figure 2. Shows what we expected. Our expectation of the experiment’s outcome was based on the results seen on plaque assays. During phage proliferation, plaques were seen Figure 8
Although bacteria grew (Figure 3) we still could not analyze our results due to our limited ability to observe the phages. Nevertheless, we could make models approximate what would happen if the bacteria had grown in the expected manner. This is presented by Bio render. Figure 4, Figure 5.
Figures 4 and 5 show what we think would have happened if one of them occurred. Figure 4 demonstrates the Null hypothesis.
The Subsequent figures demonstrate the work done and the trials we are currently running to improve the experiment. (Figure 6, Figure 7, Figure 8)
Source of error
This experiment, although it could not demonstrate the phage dynamics, the renditions provide crucial information on bacteriophage activity. However, a virtual rendition is minimal compared to what a test could provide. Therefore, to convey results, one must speak on sources of errors to which this test was certainly exposed. The foremost error that hindered this test was the incubator’s predicament. If the incubator had worked or if the bacteria were kept at an appropriate temperature of 25-30 degrees Celsius.
Another significant source of error is human error such as inconsistency or inaccurate measurement. In addition to inaccuracies in measuring liquids. Concerning the phages and human error plaque extraction/picking could have been done ineffectively, thus hindering the overall phage concentration nevertheless we held a preliminary test for the phages where we found them able to grow in visible plaques, then we extracted them. While there is a high possibility of human error, our team took great care to minimize errors arising from human measurement and laboratory techniques.
Further areas of mismanagement could have stemmed from bacteriophage denaturing due to temperature inconsistencies. However, we did our best to keep the temperature appropriate for the phages, although the incubator was not able to hose them adequately. Other than that, during the procedure, the temperature gun and water bath were used to keep liquid solutions at the correct temperature. Therefore, this source of error must be minimal. Other phage errors include pH; the pH could affect the protein structure which could denature the portions or make them less effective.
Moving on from phages to bacteria, it is important to look at natural variation of bacteria, but also in phages as a source of error is possible. Natural variation of bacteriophage infectivity could influence the experiment and distribution. For example, were phages confined to the bottom of the plate or to a specific side? Further natural variations include the infectivity ratio. The infectivity ratio of viruses differs by species and could be affected by variation of the ratio. In addition, natural variation in bacteria is seen in differences in growth and consumption of resources. However natural variation is primarily attributed to evolutionary dynamics. Mutation and evolution of the bacteria are also linked to natural variation, but they can be chiefly controlled by the plate dimensions. It is possible that different dimensions of the Pyrex tray plate could yield different evolutionary dynamics and illustrate a differential result regarding bacteriophage resistance. A wider front would increase the effective population size and thus the mutation probability, while a longer run between strips between steps would increase the natural selection among lineages. To observe this a plate would have to be designed. However, to keep everything as equal as possible a square 8-inch by 8-inch Pyrex tray was used.
Finally, one of the biggest sources of error was contamination. We attempted to be as sterile as possible by autoclaving agar materials and equipment. However, not all materials could be autoclaved such as the Pyrex tray. The agar at one point had to be touched with clean gloves during the pouring step thus contaminating it. To mitigate the contact, we will change the method of pouring agar plate. Rather than pouring agar mixtures onto plate horizontally we will change the way of pouring it. We are currently exploring measures such as layering agar horizontally in a beaker then placing it on a tray without touching the agar. Lastly, we intend to set up positive controls in a different manner, rather than having controls be on the Pyrex tray, we will have them on separate Petri dishes. We will still focus on phages as this method’s uniqueness focuses on that.
Moving onwards, since this experiment was not able to convey proper analysis of the effects of bacteria phages, we are now trying to optimize our methods. First, we include more of Harvard’s methodology in our test, such as dying the agar. The Hardvard experiment used India ink to facilitate analysis by providing contrast. We tested out growing bacteria with dark blue food colorant to test for contrast and wither the dye was toxic to the bacteria(F6). Secondly, we developed a new method of pouring agar without much contamination. This method involves layering agar vertically rather than horizontally.
Other adjustments to this test would be using E. coli b instead of K12, there have been experiments using k12 some studies show that E. coli b is more susceptible to the pages, and this susceptibility could have shown results. Additionally, a known concentration of bacteria in the plate for quantitative analysis and propagating phages in liquid media will be done to propagate phages in greater concentrations rather than in plaques. The test must be ameliorated. Despite that, this experiment emphasizes the value of carrying out research with a strict and rigorous experimental design, while solving problems in the pursuit of knowledge.
References
Adedeji, W. A. (2016, December). The treasure is called antibiotics. Annals of Ibadan postgraduate medicine. Retrieved December 17, 2022, from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5354621/#:~:text=The%20average%20life%20expectancy%20at,were%20rampant
ADS; L. N. A. C. (2018, January 20). Horizontal transfer of antibiotic resistance genes in clinical environments. Canadian journal of microbiology. Retrieved December 9, 2022, from https://pubmed.ncbi.nlm.nih.gov/30248271/
Baym, M., Lieberman, T. D., Kelsic, E. D., Chait, R., Gross, R., Yelin, I., & Kishony, R. (2016). Spatiotemporal microbial evolution on antibiotic landscapes. Science (New York, N.Y.), 353(6304), 1147–1151. https://doi.org/10.1126/science.aag0822
Brown, N. (2015, October). (PDF) antibiotic resistance: What, why, where when and how? – researchgate. researchgate. Retrieved February 28, 2023, from https://www.researchgate.net/publication/283211266_Antibiotic_resistance_What_why_where_when_and_how
Caddick, A. (2016, October 4). The overuse of antimicrobial agents in agriculture. Open Access Government. Retrieved November 14, 2022, from https://www.openaccessgovernment.org/overuse-antimicrobial-agents-agriculture/29045/
Dedrick, R. M., Smith, B. E., Cristinziano, M., Freeman, K. G., Belessis, Y., Whitney Brown, A., Cohen, K. A., Davidson, R. M., van Duin, D., Gainey, A., Garcia, C. B., Robert George, C. R., Haidar, G., Ip, W., Iredell, J., Khatami, A., Little, J. S., Malmivaara, K., McMullan, B. J., . . . Hatfull, G. F. (2023). Phage Therapy of Mycobacterium Infections: Compassionate Use of Phages in 20 Patients with Drug-Resistant Mycobacterial Disease. Clinical Infectious Diseases, 76(1), 103-112. https://doi.org/10.1093/cid/ciac453
Enoch, D. A. (2015, October 15). (PDF) antibiotic resistance: What, why, where when and how? – researchgate. https://www.researchgate.net. Retrieved December 8, 2022, from https://www.researchgate.net/publication/283211266_Antibiotic_resistance_What_why_where_when_and_how
Furfaro, L. L., Payne, M. S., & Chang, B. J. (2018, October 5). Bacteriophage therapy: Clinical trials and regulatory hurdles. Frontiers. Retrieved December 7, 2022, from https://www.frontiersin.org/articles/10.3389/fcimb.2018.00376/full
Hancock, R. E. W., & Reeves, P. (1975, August 6). Home – PMC – NCBI. National Center for Biotechnology Information. Retrieved March 8, 2023, from https://www.ncbi.nlm.nih.gov/pmc/
Ike, Y., Pantosti, A., Noble, W. C., Werner, G., Schouten, M. A., Arthur, M., Moore, P. R., Luecke, W., Jukes, T. H., Witte, W., Levy, S. B., Linton, A. H., Hummel, R., Tschäpe, H., Casadewall, B., Klare, I., & Chadwick, R. P. (2000, December 21). Selective pressure by antibiotic use in livestock. International Journal of Antimicrobial Agents. Retrieved December 10, 2022, from https://www.sciencedirect.com/science/article/pii/S0924857900003010
Jia, Y., Wang, Z., Fang, D., Yang, B., Li, R., Liu, Y., & AbstractThe emergence and increasing prevalence of antibiotic resistance pose a global public risk for human health. (2021, April 7). Acetaminophen promotes horizontal transfer of plasmid-borne multiple antibiotic resistance genes. Science of The Total Environment. Retrieved December 28, 2022, from https://www.sciencedirect.com/science/article/abs/pii/S0048969721019860
Kasman , L. M. (2022). Bacteriophages – statpearls – NCBI bookshelf. NIH.gov . https://www.ncbi.nlm.nih.gov/books/NBK493185/
Khan, L. B. (2018, August 31). American Society for Microbiology. journals.asm.org. Retrieved February 28, 2023, from https://journals.asm.org/doi/10.1128/jmbe.v19i2.1542
Landecker, H. (2016). Antibiotic resistance and the biology of history – sage journals. Body and Society. Retrieved December 7, 2022, from https://journals.sagepub.com/doi/10.1177/1357034X14561341
Lee, A. (2020, April 15). Plaque assay. Plaque Assay. Retrieved December 8, 2022, from https://www.protocols.io/view/plaque-assay-be6sjhee.html#:~:text=Bacteriophages%20(phage)%20are%20viruses%20that,concentration%20of%20a%20given%20sample.
Levy, S. B. (n.d.). The challenge of antibiotic resistance – hummingbirds.arizona.edu. http://www.hummingbirds.arizona.edu/. Retrieved December 18, 2022, from http://www.hummingbirds.arizona.edu/Courses/Ecol409_509/levy.pdf
Pincus, N. B., Reckhow, J. D., Saleem, D., Jammeh, M. L., Datta, S. K., & Myles, I. A. (2015). Strain Specific Phage Treatment for Staphylococcus aureus Infection Is Influenced by Host Immunity and Site of Infection. PLOS ONE, 10(4), e0124280. https://doi.org/10.1371/journal.pone.0124280
Liu Y;Tong Z;Shi J;Jia Y;Yang K;Wang Z; L. (2020, August 8). Correlation between exogenous compounds and the horizontal transfer of plasmid-borne antibiotic resistance genes. Microorganisms. Retrieved November 10, 2022, from https://pubmed.ncbi.nlm.nih.gov/32784449/
Taylor, P. (2020, July). A new study reveals the use of antibiotics on crops is more widespread than previously thought. CABI.org. Retrieved November 21, 2022, from https://www.cabi.org/news-article/new-study-reveals-use-of-antibiotics-on-crops-is-more-widespread-than-previously-thought/
USA, C. D. C. (2023, February 7). FastStats – life expectancy. Centers for Disease Control and Prevention. Retrieved February 26, 2023, from https://www.cdc.gov/nchs/fastats/life-expectancy.htm
Van Boekel, R. (2015, February 18). Global trends in antimicrobial use in food animals | PNAS. Global trends in antimicrobial use in food animals. Retrieved March 24, 2023, from https://www.pnas.org/doi/10.1073/pnas.1503141112