Pierre Collet, Year 2 Research
Abstract
Since the development of the first antibiotic, Penicillin, in 1928, scientists have identified the possible risk of antibiotic resistance. Because bacteria are living organisms, they can evolve and adapt to current treatment methods, causing the creation of superbugs. These antibiotic resistant bacteria are much more difficult to treat and are predicted to cause millions of deaths and trillions of dollars in the years to come. In this experiment, several generations of the bacteria Pseudomonas fluorescens were created, compared, and tested for antibiotic resistance to Neomycin. It was found that Pseudomonas fluorescens became antibiotic resistant after only one generation of being exposed to the Neomycin.
Introduction
Antibiotic resistance occurs when a microbe resists the effects of medication that once could successfully eliminate it. This type of bacteria has grown immune to traditional antibiotics through random mutation. An Example is Neisseria gonorrhoeae. In the past, azithromycin, cefixime, and ceftriaxone have been used to treat it; however, strands of Gonorrhea are starting to become resistant, causing cases of Gonorrhea in Australia to be reported as immune to most antibiotics (Australian Government Department of Health, 2018).
Antimicrobial Resistance occurs by mutation. During DNA replication, random mutations can arise by chance, leading to different traits. Suppose a spontaneous mutation in a bacterium causes it to be resistant to antibiotic X. In that case, all the non-resistant strains will die when exposed to antibiotic X, leaving the resistant ones to reproduce. This means, through the antibiotic’s selection, we have created a form of bacteria resistant to the treatment.
Antibiotic Resistance is becoming more and more prevalent as the use of antibiotics increases. According to the WHO, in late 2014, antibiotic-resistant organisms are predicted to cause 10 million deaths per year and cost 100 trillion dollars by 2050 (World Health Organization, 2020). This is something that we would have to learn to live with. The rise of antibiotic resistance can largely be attributed to the agricultural industry and the medical practice of overprescription. According to the FDA, more than 20 million pounds of antibiotic drugs were sold for use on livestock farms in 2014, making 80% of the antibiotics sold (FDA, 2015). In addition, the center for disease control and prevention stated that up to one-half of antibiotic use in humans is unnecessary and inappropriate (CDC, 2016)
The extreme prescription rates of antibiotics has lead researcher Marin Kollef from the University of Washington St. Louis, among other professionals, to heavily criticize the practice. Kollef attributes high antibiotic prescription rates to “spiralling empiricism”, explaining that medical professionals often prescribe antibiotics when in doubt instead of trying alternatives (Kollef et al. 1994).
Patients diagnosed with antibiotic resistance are found to have higher rates of mortality and are a greater cost to the medical system. For instance, “Ventilator-associated pneumonia due to antibiotic-resistant bacteria has become recognized as an important problem (Kollef et al. 1994).
This study seeks to discover the number of generations needed to create antibiotic-resistant bacteria in a lab environment. Precisely, the goal is to treat a common bacteria with an antibiotic that is “one of the most effective drugs against it” and discover the number of generations necessary for that antibiotic to have a mitigatory effect (Waisbren et al. 1958). If the antibiotic is no longer able to visibly eliminate the bacteria, the experiment will have succeeded in creating antibiotic-resistant bacteria.
Materials And Methods
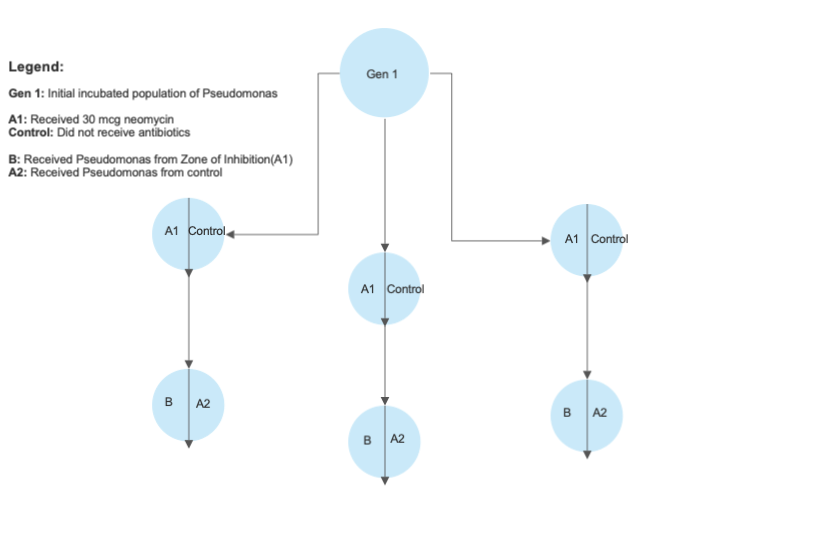
A strand of Pseudomonas fluorescens was cultivated in nutrient agar for one generation inside an incubator at 37 degrees celsius. The incubation period would last 1 week, creating the generation 1 Pseudomonas. The generation 1 Pseudomonas was then plated into 6 distinct sections of 3 nutrient agar plates. The agar sections were split into two groups, with Group A1 receiving a 30mcg disc of neomycin and Group Control receiving no antibiotics. The plated bacteria was then placed into the same incubator at the same temperature and grown for another week, creating the generation 2 Pseudomonas.
The generation 2 Pseudomonas was then measured with a standard ruler for the size of the zone of inhibition around the neomycin. As the radius was not constant at all points, the largest radius was taken as the measurement.
For generation 3, The specimens of bacteria found within the zone of inhibition of neomycin would be collected and placed into 3 new plates of nutrient agar. For control purposes, each agar plate was cut in half. One side (Group B) received bacteria from the zone of inhibition, and the other side (Group A2) received bacteria from the non-treated sections of Pseudomonas. 30mcg discs of neomycin were then placed on both halves of the plates, and the Pseudomonas was left to incubate for a week, creating the generation 3 Pseudomonas
Finally, the zones of inhibitions around the generation 3 Pseudomonas were measured with a standard 30 cm ruler. If the radius was less than 1mm, the bacteria was defined as resistant.
Results
Table 1: Measurement of the zone of inhibitions across 6 samples in 2 weeks.
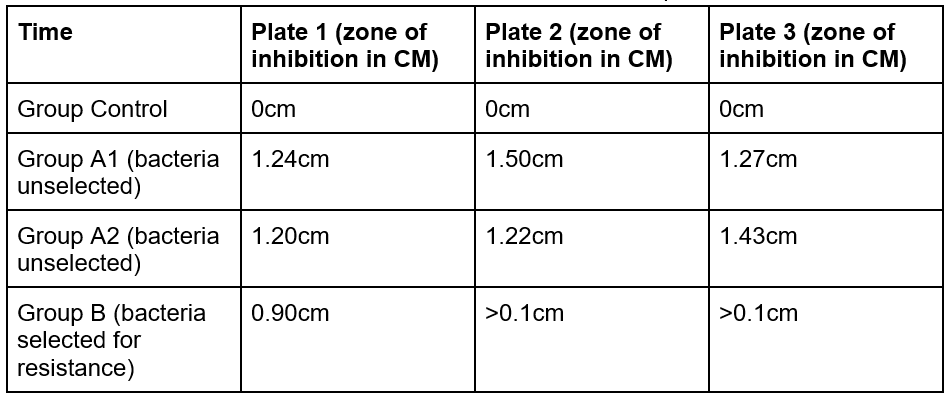
There was a clear distinction between the size of inhibition for the experimental Group B compared to the rest. While in plates 2 and 3, the antibiotic resistant Group B had a zone of inhibition less than 0.1 CM, Group A1 and A2 had values greater than 1 CM for all Plates. Furthermore, the largest value of Group B is 25% smaller than the smallest value in Group A2 and 27% than that of Group A1.
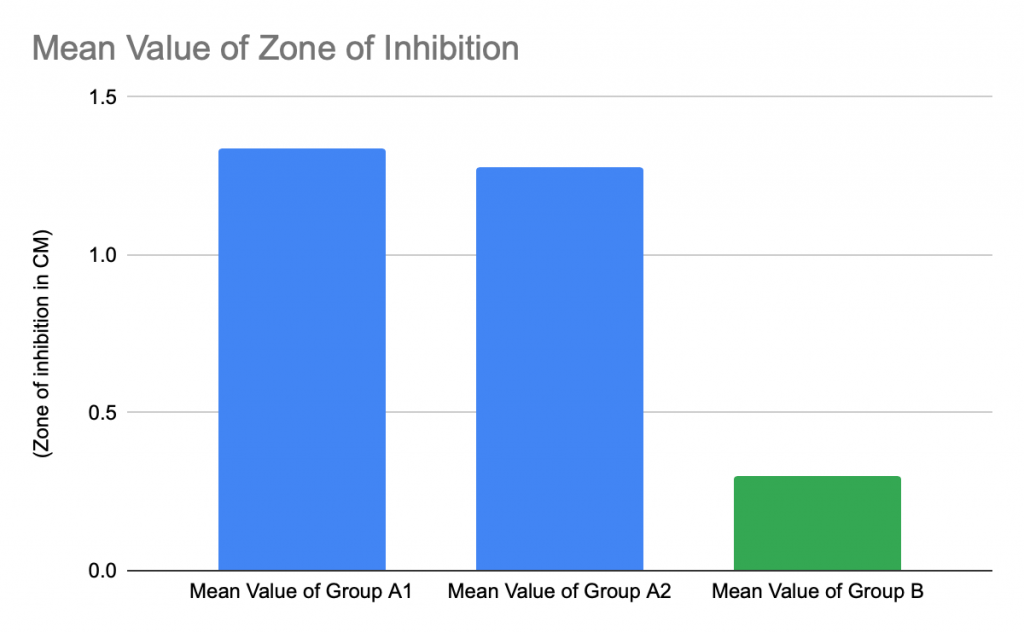
Figure 1: Bar Graph representing average value of experimental and control groups
The selection of antibiotic resistant bacteria within the zone of inhibition led to a much smaller zone of inhibition in Group B when exposed to the same 30mcg tablet of Neomycin than Group A. The mean value of Group B is 88% less than the mean value of Group A1 and 87% less than the mean value of Group A2. That being said, Group A1 and Group A2 have very similar mean values that differentiate by less than 0.06 CM.
Table 2: Qualitative observations of the zone of inhibitions across 6 samples
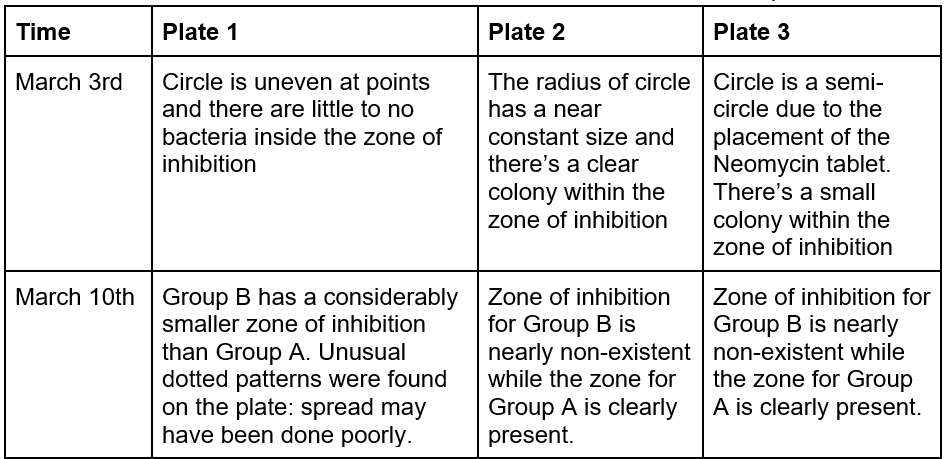
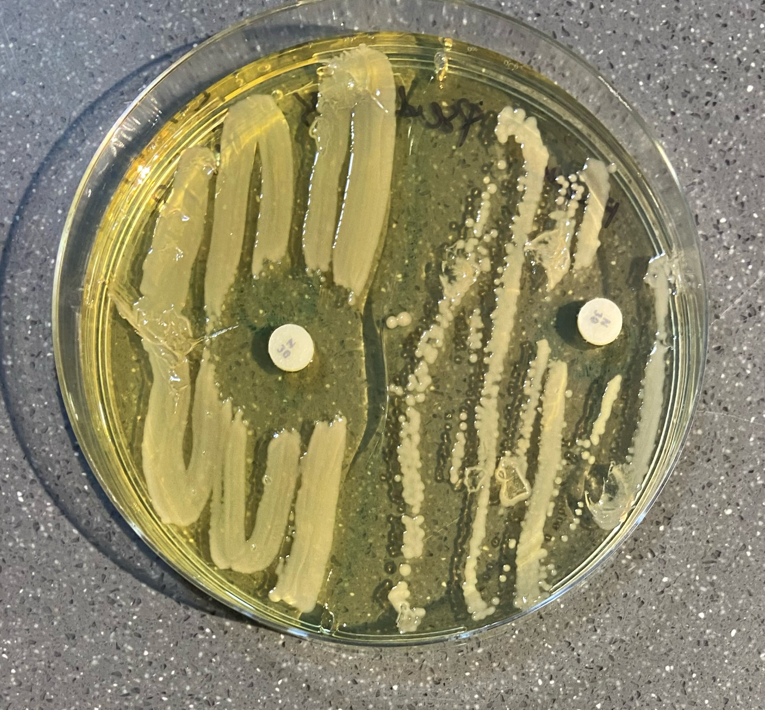
Figure 2: Unusual dotted pattern in lower half of Group B, plate 1.
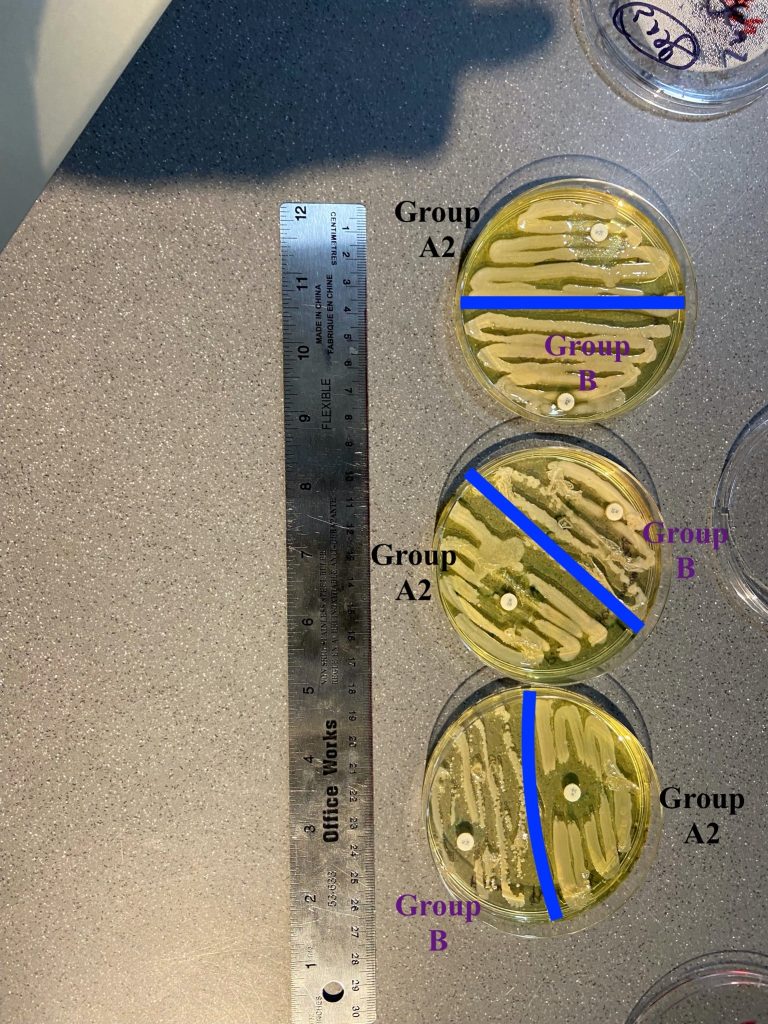
Figure 3: measurements of Group B and A2
Discussion
Within one generation of selective breeding, the effectiveness of Neomycin on Pseudomonas was greatly reduced. This means, at least in a controlled environment, bacteria are able to readily adapt to antibiotics. Although antibiotic resistance in this experiment was selected for, it can still most certainly be replicated in nature. All it takes is for a single resistant bacteria to make its way into a nutrient rich environment for it to spread. For instance, “it would take only 10 generations for that single cell to grow into a colony of more than 1,000, and just 10 more generations for it to erupt into a colony of more than 1 million” (Pray, 2008). Furthermore, since some bacteria are able to duplicate every 30 minutes, it would only take a matter of hours before colonies of millions form.
That being said, It’s important to note that this study was conducted in a very supervised environment, the bacteria had access to a safe incubated and nutrient rich environment to grow and thrive in. If this study was conducted in an open environment, the time it would take to reach the same level of resistance would probably be significantly larger.
The results were generally consistent with my predictions. It made sense that Group B had the smallest zone of inhibition, since the bacteria was actively selected for an ability to survive within the zone of inhibition of Neomycin in Group A1. Furthermore, it was expected that Group A1 and A2 had similar size zones as they were both only exposed to 1 tablet of Neomycin and the bacteria was not selected for. Group B in Plate 1, on the other hand, had an unexpected result. While its counterparts in Plate 2 and 3 had a zone of inhibition of less than 0.1 CM, it had a value of 0.9 CM. That being said, When looking at the qualitative analysis, Plate 1 had Pseudomonas with an usual dotted pattern (Figure 2). This could possibly explain the outlier, perhaps the bacteria was improperly plated.
What can be done to help with the rise of antibiotic resistance? Every year, the agricultural and medical industry use millions of tons of antibiotics when they’re not necessary. (CDC, 2016) In response, the government should make legislation to ban the use of antibiotics on livestock as it is done in europe. Furthermore, novel antibiotics should be protected for the most extreme cases possible, because we don’t want antibiotics to grow resistant to them. Drops in antibiotic usage means that bacteria that have developed resistance to antibiotics through random mutation are less likely to be selected by natural selection. (FDA, 2019)
References
“Antimicrobial Resistance.” Cambridge Infectious Diseases, 14 Aug. 2019, www.infectiousdisease.cam.ac.uk/research/active-research-projects/bacterial-pathogens.
Sample, Ian. “Powerful Antibiotic Discovered Using Machine Learning for First Time.” The Guardian, Guardian News and Media, 20 Feb. 2020, www.theguardian.com/society/2020/feb/20/antibiotic-that-kills-drug-resistant-bacteria-discovered-through-ai.
Society, Microbiology. “The History of Antibiotics.” Microbiology Society, microbiologysociety.org/members-outreach-resources/outreach-resources/antibiotics-unearthed/antibiotics-and-antibiotic-resistance/the-history-of-antibiotics.html.
“Superbug: Definition of Superbug by Oxford Dictionary on Lexico.com Also Meaning of Superbug.” Lexico Dictionaries | English, Lexico Dictionaries, www.lexico.com/en/definition/superbug.
“Cartoon Depicts Superbug Microorganism.” Shutterstock, 15 June 2020, image.shutterstock.com/image-vector/cartoon-depicts-super-bug-microorganism-260nw-703941442.jpg.
“Checklist Box.” Kaislerg, 20 June 2020, www.kaiserlg.com/wp-content/uploads/2018/10/checklist-box-PTH8EMY.jpg.
Cow. 13 June 2020, encrypted-tbn0.gstatic.com/images?q=tbn%3AANd9GcRv9Ahq9vZNdGCS4F6SrdyPixEfB1SzRLP3lg&usqp=CAU.
“One Hundred Dollar Bill Medical.” Shutterstock, 13 June 2020, image.shutterstock.com/image-photo/one-hundred-dollar-bill-medical-600w-1664816185.jpg.
“Antibacterial Resistance.” Medicalnewstoday, 14 June 2020, post.medicalnewstoday.com/wp-content/uploads/sites/3/2020/01/327093_2200-1200×628.jpg.
“Overusing Antibiotics.” Healthywomen, 20 June 2020, www.healthywomen.org/sites/default/files/overusing-antibiotics_1.jpg.
“Pills.” Theconversation, 13 June 2020, images.theconversation.com/files/313959/original/file-20200206-43084-f2m67.jpg?ixlib=rb-1.1.0&rect=0%2C23%2C5184%2C3370&q=45&auto=format&w=926&fit=clip.
“MRSA in Petri Dish.” Medicalnewstoday, 15 June 2020, i0.wp.com/cdn-prod.medicalnewstoday.com/content/images/articles/327/327050/mrsa-in-petri-dish.jpg?w=1155&h=1541.
“Virus Scientists.” Nyt, static01.nyt.com/images/2020/03/31/world/00virus-scientists1/00virus-scientists1-mediumSquareAt3X-v2.jpg.
“Antibiotics – Neomycin, Polymyxin B, Bacitracin and Tyrothricin: Nejm.” New England Journal of Medicine, https://www.nejm.org/doi/full/10.1056/NEJM195806122582409.
Commissioner, Office of the. “Combating Antibiotic Resistance.” U.S. Food and Drug Administration, FDA, https://www.fda.gov/consumers/consumer-updates/combating-antibiotic-resistance.